symfer¶
symfer is a tool suite, written mostly in Python, for performing probabilistic inference: calculating arbitrary probabilities like 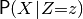 , given a model
, given a model 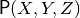 and observation
and observation 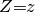 . A script for this specific query is constructed during a symbolic phase in Python, and executed in a separate numeric phase (possibly on a different platform/machine).
. A script for this specific query is constructed during a symbolic phase in Python, and executed in a separate numeric phase (possibly on a different platform/machine).
Basic usage¶
From the Python prompt, load a Bayesian network model (stored in Hugin format):
>>> import symfer as s
>>> model = s.loadhugin('extended-student.net')
Select a query variable 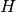 and set observation
and set observation 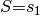 :
:
>>> query = ['H']
>>> obs = {'S':'s1'}
Symbolic phase: construct an inference script (here using variable elimination with minweight heuristic).
>>> ve = s.ve_minweight(model,query)
>>> ve_obs = s.indextree(obs,ve)
Note
It is easy to tweak the supplied Inference algorithms or even roll your own.
Numeric phase: Execute the script, here using the internal evaluator.
>>> s.evaluate(ve_obs)
Multinom(dom=[{'H': ['h0', 'h1']}],par=[0.0677442825, 0.20725571750000002])
This result represents the distribution 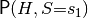 .
.
Note
Instead of directly executing the script from the same Python interpreter, it can also be evaluated at a different machine and/or using Java/C. See Evaluation options.
Tutorial¶
Reference¶
License¶
symfer is licensed under the 2-clause BSD license.
If you use it in published research, please refer to the following article: [pdf]
Sander Evers and Peter J.F. Lucas, A framework for development, teaching and deployment of inference algorithms. In: Andrés Cano, Manuel Gómez-Olmedo, Thomas D. Nielsen (Eds.), PGM 2012, Sixth European workshop on Probabilistic Graphical Models, Electronic proceedings, pp. 99–106, http://leo.ugr.es/pgm2012/proceedings/proceedings.pdf
Development¶
Currently, symfer has only one developer: Sander Evers. However, he is happy to accept any contributions, or team up with you if you’d like to share some responsibility. Reach him at sandr {at} dds {dot} nl.
Acknowledgements¶
This software has evolved from a research tool developed at the University of Twente (DB group) and Radboud University Nijmegen (MBSD group).

